Description
Introduction
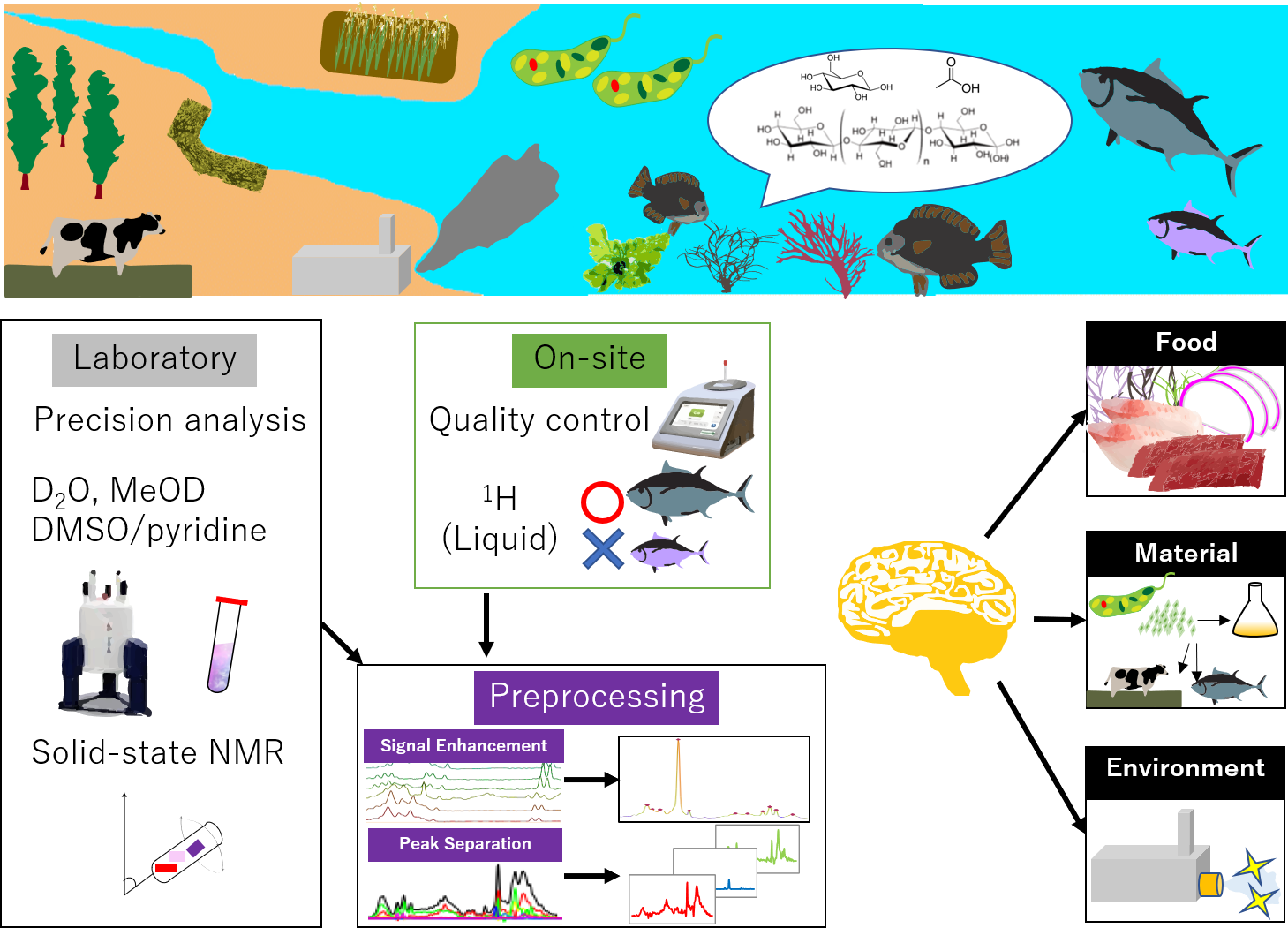
Environmental problems such as marine pollution, destruction of land and fresh water ecosystems, depletion of resources including energy, raw materials, and food, and health problems are some of the global challenges of modern society. The realization of a materials-circulating society, including use of renewable energy and production of sustainable food and materials, is increasingly important. With the rapid development of information and communication technology in recent years, it is expected that innovations in environmental science, sustainable resources, materials, foods, and medicine, will be integrated by effectively connecting the accumulating scientific data and real-world information. As a result, digital innovations in the analyses of natural mixtures, such as biogeochemical samples from the environment and molecular complexities from biological tissues, are becoming important both for a sustainable society and for healthcare.
Nuclear magnetic resonance (NMR) approaches to natural mixture analysis are being developed as a strategy to evaluate homeostatic stages via molecular compositional changes in healthcare, foods, natural materials, biomass utilizations, and environmental ecology. Alongside, there have been many advances in NMR technology, including high-field NMR over 1 GHz using high-temperature superconducting materials, hyperpolarization and photodetection NMR using diamond nitrogen-vacancy centers, zero-magnetic field NMR, and compact and benchtop NMR instruments that have become highly cost-effective owing to the marked progress in permanent magnet materials. These innovations in NMR hardware are likely to be applied not only to precise analysis by high magnetic field NMR in the laboratory but also to homeostatic assessments of environment and health, and quality control in the fields of agriculture, forestry, and fishery.
Thus, identification of molecules contained in mixtures is an important task in NMR analysis. Because the physical and chemical properties of these molecules can be extremely diverse, various sample preparation methods and pulse sequences have been used for mixture analysis. Depending on the target molecules under analysis, the sample preparation method may range from solid-state to polar and nonpolar solvent systems. When targeting small molecules, for example, solution NMR in a polar or semipolar solvent system such as deuterated water (D2O) or deuterated methanol (MeOD) is generally used. On the other hand, macromolecules can be evaluated by using a dimethyl sulfoxide (DMSO) solubilized system or by solid-state 13C- cross-polarization magic-angle spinning (CP-MAS) NMR. 1D-NMR and 2D-NMR such as 1H-13C hetero-nuclear single quantum coherence (HSQC) and 2D-1H-J resolved (2D-Jres) spectroscopy are also useful for applications where stable isotope labeling experiments cannot be applied.
Nevertheless, such molecular assignments remain difficult owing to the problems of spectral overlap, and a lack of available reference spectra or convenient molecular assignment tools specific to the molecules and conditions of interest. Databases and analytical tools for traditional major metabolomics studies such as HMDB, BMRB, BML-NMR, MMCD, NMRShiftDB, TOCCATA, COLMAR, MetaboLights, MetaboAnalyst, SpinAssign, and SpinCouple focus on the analysis of low molecular mass metabolites by high magnetic field solution NMR. For the analysis of macromolecular mixtures derived from environmental samples and living organisms, however, solid-state CP-MAS spectral can characterize insoluble samples, whereas HSQC spectral data in DMSO solvent are required to characterize soluble samples. The BMRB contains reference NMR data on biomolecules in various solvents such as DMSO and methanol, but it is limited to partial structural data for polysaccharides. Although Bm-Char of ECOMICS can be used to characterize chemical structures from the HSQC spectrum of a biomass sample, there remain insufficient databases and analytical tools for complex mixtures of similarly structured macromolecules, or for solid CP-MAS NMR, which has typically very low resolution, or low-field benchtop NMR.
Database and Web-tools
In order to overcome these problems, here we have developed InterSpin, an integrated supportive webtool comprising freely accessible preprocessing tools, a database, and molecular assignment webtools to advance signal assignment in low- and high-field NMR analyses of small- to macro-molecular mixtures.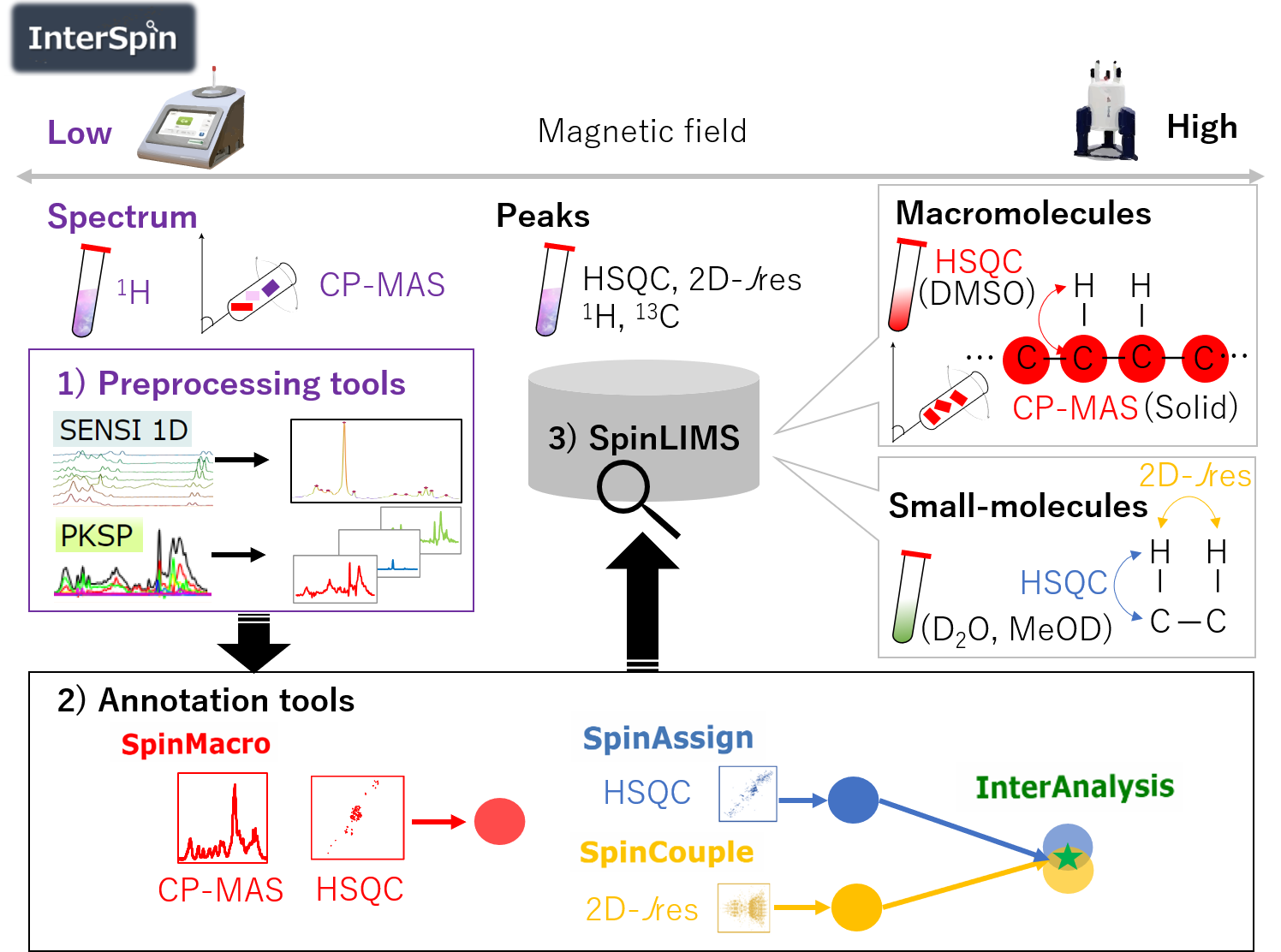
InterSpin comprises the following three elements.
1) Spectrum preprocessing tools. In the case of a broad spectrum obtained from low-field benchtop 1H-NMR or solid-state 13C-CP-MAS, SENSI (SENsitivity improvement with Spectral Integration) overcomes the problem of low signal-to-noise ratio by increasing resolution through the integration of multiple spectra, while PKSP performs effective peak separation by a multivariate spectral decomposition method.
2) Molecular assignment tools. SpinMacro simplifies the macromolecular assignment of a solid CP-MAS spectrum or a DMSO-solubilized 1H-13C HSQC spectrum. SpinAssign searches the SpinLIMS database for a compound corresponding to the HSQC NMR peaks. SpinCouple can assign 1H-J 2D-Jres NMR peaks. InterAnalysis is a Venn diagram-type highly accurate annotation tool that narrows down candidate molecules by using correlation peaks from both the HSQC spectrum and the 2D-Jres spectrum. In the bottom right of the figure, blue, yellow, and red circles represent a set of search results; the green star represents the narrowed down set.
3) SpinLIMS (InterSpin Laboratory Information Management System) database. The database includes reference solid-state CP-MAS spectra and solution-state HSQC spectra (DMSO) for macromolecules, and reference solution-state HSQC and 2D-Jres spectra (D2O and MeOD) for small molecules.

